Part:BBa_K4274030
ptac-RiboJ-B0034-ClSS_S533A-B0034-ERG20-B0015
The expression cassette ptac-RiboJ-B0034-ClSS_S533A-B0034-ERG20-B0015 includes ClSS_S533A (Part: BBa_K4274000) and ERG20 (Part: BBa_K849001). It is used to heterologously express santalene synthase and farnesyl pyrophosphate synthase in E. coli strain DH5a (tnaA-), which take part in the alpha-santalene production pathways of IPP ⇄DMAPP → GPP → FPP → alpha-santalene. This part is also composed of the IPTG-inducible promoter ptac-RiboJ (Part: BBa_K3552015), the ribosomal binding site B0034 (Part: BBa_B0034), and the strong terminator B0015 (Part: BBa_B0015).
To optimize the production of alpha-santalene, the part collection we built includes: Parts to optimize the activity of farnesyl pyrophosphate synthase, ERG20_F96W (Part: BBa_K4274002) and ptac-RiboJ-B0034-ClSS_S533A-B0034-ERG20_F96W-B0015 (Part: BBa_K4274031). Parts to optimize the seceation of santalene synthase, CmR29M2-ClSS_S533A (Part: BBa_K4274001) and ptac-RiboJ-B0034-CmR29M2-ClSS_S533A-B0034-ERG20_F96W-B0015 (Part: BBa_K4274032). Parts expressing fusion protein ClSS_S533A-FL-ERG20_F96W BBa_K4274003 and BBa_K4274033. Our part collection can be used to help and inspire future teams to design and perfect different alpha-santalene production pathways in E. coli.
Usage and Biology
ClSS_S533A is an optimized biobrick part encoding the gene for alpha-santalene synthase. The enzyme could catalyze the conversion of the common isoprenoid intermediate farnesyl pyrophosphate (FPP) into the alpha-santelene in a single step. ERG20 is a part encoding farnesyl pyrophosphate synthase producing FPP. In our cases, ClSS_S533A and ERG20 could be heterologously expressed in E.coli under the control of ptac-RiboJ promoter and B0015 terminator.
Source
ClSS_S533A (Part: BBa_K4274000) is from Clausena lansium, and ERG20 (Part: BBa_K849001) is from S. cerevisiae.
Characterization
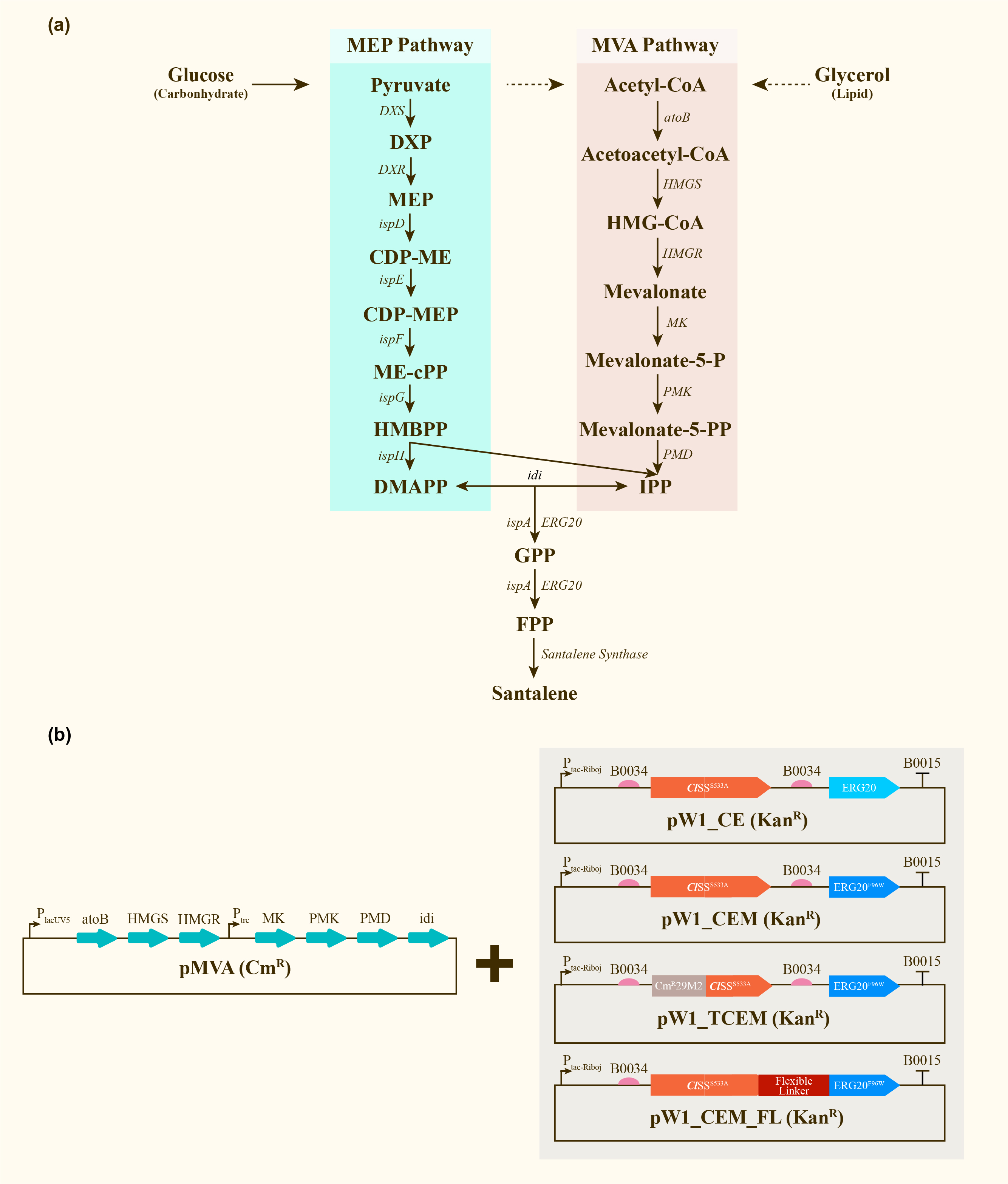
After engineering, E. coli could utilize both MEP pathway and MVA pathway for the universal precursors isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP), then synthesize santalene with the help of FPP Synthase (FPPS) and santalene synthase (SS). Except heterologously expressed MVA pathway and ERG20 of Saccharomyces cerevisiae and santalene synthase of Clausena lansium (ClSS), several modifications upon ERG20 or ClSS by amino acid mutation, binding to a hydrophillic tag and the construction of fusion protein were tested for the higher yield of santalene. Therefore, with the help of the co-transformation of pMVA plasmid with various pW1 plasmids, including pW1_CE, pW1_CEM, pW1_TCEM and pW1_CEM_FL, different strains like CE, CEM, TCEM, CEM_FL were successfully constructed (Figure 1). The complete pathway we designed for producing santalene in E. coli is illustrated in Figure 1.
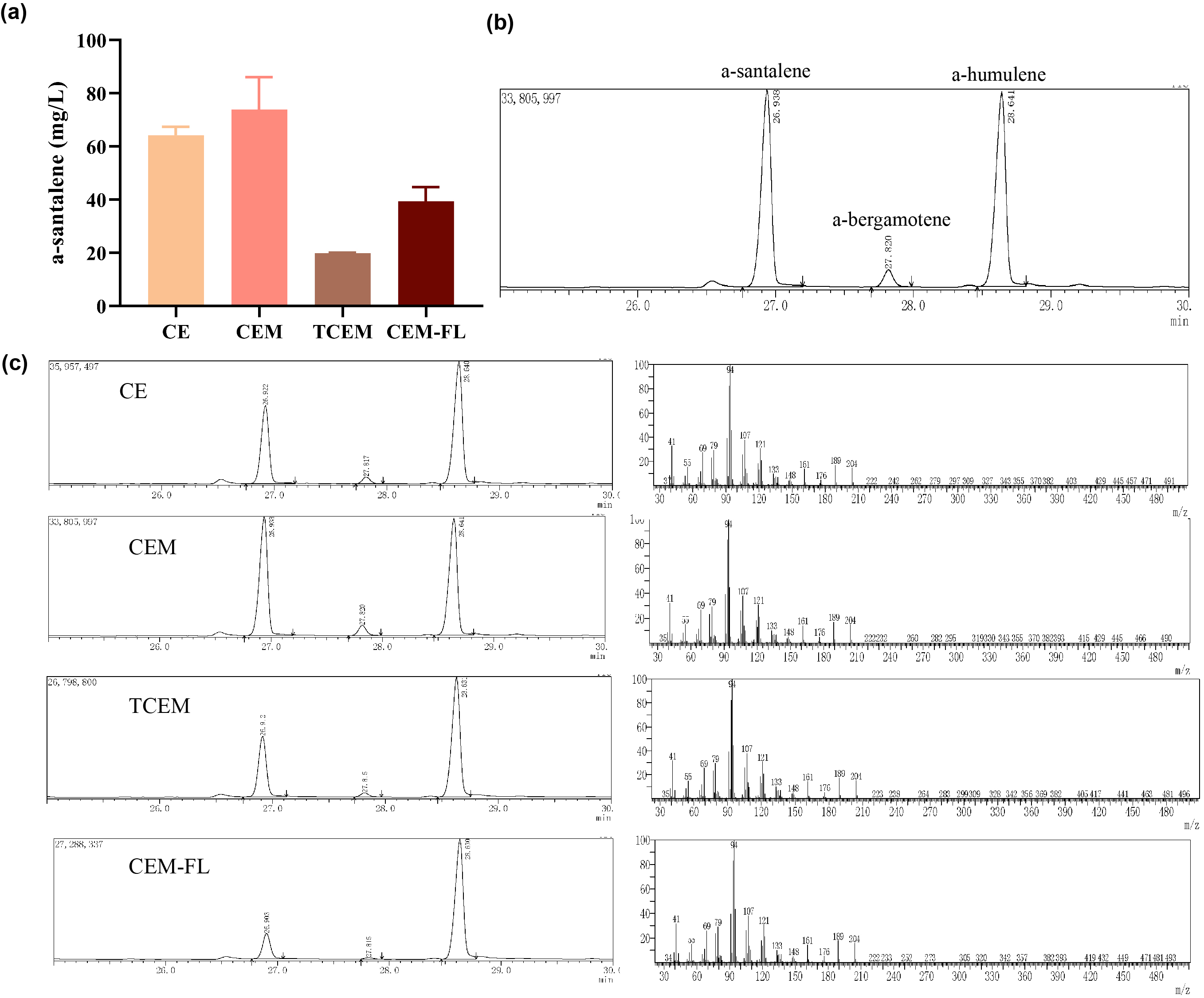
Afterwards, the various engineering of E.coli DH5α ∆TnaA mentioned above were used for santalene production. After rapid centrifugation, the supernatant of dodecane was spiked with with 0.475 g/L a-humulene as an internal standard, and then injected into GC/MS for verification of α-santalene production. It turned out that all samples from four strains appeared a significant peak at the retention time of 26-27 min, and various peak area of different samples exhibited santalene production with differing levels, indicating the general success of E. coli engineering. It can be concluded that the E. coli strain CEM (with pW1_CEM plasmid) produces the maximal level of α-santalene compared to other strains (73.93 mg/L). Furthermore, our study elucidates that the mutation of 96th amino acid into tryptophan could increase the yield of α-santalene by about 20%, substantiating the prominent performance of ERG20F96W in enhancing the supply of FPP and α-santalene production in E. coli (Figure 2).
Sequence and features
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 1541
- 1000COMPATIBLE WITH RFC[1000]
| None |
